Original Article
Molecular Characterization of Staphylococcus aureus Isolates Obtained from Hemodialyzed
Patients at the Hospital de Clínicas of Paraguay: A pilot study
Rubén Arturo Silvero-Isidre1, Fátima Rodríguez-Acosta1, César Rodrigo Cristaldo-Vargas1, Genaro Américo Velázquez-Romero1, José Félix Plans-Perrota1, Rosa María Guillén-Fretes1
doi: http://dx.doi.org/10.5195/ijms.2017.172
Volume 5, Number 1: 14-19
Received 09 09 2016:
Accepted 17 01 2017
ABSTRACT
Background:
Patients undergoing hemodialysis are susceptible to the nasal carriage of Staphylococcus
aureus, increasing the risk of developing infections associated with higher morbidity
and mortality. The objective of this study was to describe the frequency of S. aureus
carriage in hemodialysis patients and to perform molecular analysis of isolates by
applying multiple-locus variable analysis.
Methods:
We conducted a descriptive cross sectional study with non-probabilistic sampling that
included 28 hemodialysis patients attending the Nephrology Department of Hospital
de Clínicas in Asunción, Paraguay. We obtained clinical data from medical records
and interviews with patients. Nasal swabs were collected and analyzed by microbiological
and molecular methods.
Results:
The frequency of S. aureus carriage was 50% (14/28), 93% of which (13/14) were methicillin
resistant, 57% (6/14) were gentamicin resistant and 36% (5/14) were resistant to more
than 4 antibiotic classes. S. aureus carriers showed higher frequency of rhinitis
(p=0.02 odds ratio [OR]=6.6 (1.2-34.4)). Seven methicillin-resistant S. aureus isolates
had been analyzed by multiple-locus variable analysis, two of them showed identical
pattern bands.
Conclusion:
We found a high frequency of methicillin-resistant Staphylococcus aureus colonization
and the presence of two isolates with identical profile in the multiple-locus variable
analysis indicating the possibility of transmission between patients.
Keywords:
Staphylococcus aureus;
Hemodialysis;
Antibiotic resistance;
bacterial typing.
Introduction
S. aureus is a common pathogen causing bloodstream infections in the hospital environment.
The possibility of nasopharyngeal colonization increases the risk of endogenous infections,
and is linked to the 80% of cases of invasive S. aureus infections.1 This microorganism was frequently reported as a pathogen in patients undergoing dialysis
and kidney transplantation.2 These individuals have risk factors for colonization and infection with multidrug-resistant
S. aureus because they are exposed to frequent and prolonged use of antimicrobials.3 Furthermore, the need to use invasive devices such as catheters for venous access
are associated with a high risk of bloodstream infections.4,5 In Paraguay the data about this pathogen in hemodialysis patients is scarce and is
limited to local studies that have not been published. Other risk factors for methicillin
resistant S. aureus (MRSA) carriage are age ≥ 75 years, prolonged hospitalization,
history of repeated administration of antibiotics, type of vascular access, the frequency
of hospitalization, immunosuppressive therapy, the use of heparin in the middle of
treatment, iron overload, lack of hygiene, comorbidi-ties and proximity to other people
carrying S. aureus.6,7
Despite the great technological advances, the mortality rates in hemodialyzed (HD)
patients remain unsatisfactorily high. Along with cardiovascular disease, infections
are the leading causes of morbidity, hospitalization and mortality in this population.
The annual mortality rate for sepsis is 100 to 300 times higher in patients with end
stage renal disease than in the general population, and there is evidence of the association
between nasal carriage of MRSA and poor clinical outcome in HD out-patients.6-8
Molecular typing has been used to perform epidemiological studies at the global level.
In a specific environment over a short period of time, microorganism typing techniques
are used to study nosocomial outbreaks, local transmission, and the relationship between
carriage and infection in patients.9
The multiple-locus variable number of tandem repeat analysis (MLVA) method can be
used for the analysis of genetic variability of S. aureus isolates because it has
a high discriminatory power for the characterization of bacterial isolates, and it
is based on the variation of 7 different loci. The analysis generates multiple PCR
products that differs in size for each allele and produces a pattern of 7 bands like
a code bar. Isolates that are genetically distant present differences in their profile
of bands and those that are identical share the same pattern of bands.3,10 The objective of this pilot study was to describe the frequency of S. aureus carriage
in hemodialyzed patients attending the nephrology department of the Hospital de Clínicas,
and perform the clustering analysis of isolates by the MLVA molecular technique.
Patients and Methods
Study Design
A descriptive cross-sectional pilot study with a non-probabilistic sampling of consecutive
cases was performed on October 2013, including 28 patients undergoing hemodialysis
in the Nephrology Department of Hospital de Clínicas from Paraguay. The study was
approved by the Facultad de Ciencias Médicas Ethics Committee of the Universidad Nacional
de Asunción. Patients received information about the study and those who agreed to
participate voluntarily signed an informed consent form. For the data collection process,
we interviewed patients and revised the medical records. Two patients with missing
data concerning this study were excluded. The data collected included the patient’s
age, sex, years undergoing hemodialysis, white blood cell count, differential count
of lymphocytes, neutrophils and eosinophils expressed as number of cells/mm3, day
and turn of hemodialysis, use of antibiotics over the last six months, the use of
an invasive device, and history of rhinitis.
Collection of samples
We collected the nasal swab of dialysis patients and preserved them in Stuart medium
for transportation to the microbiology laboratory. The samples were cultured on blood
agar, chocolate agar, mannitol agar and incubated at 37°C for 24 hours. The identification
of S. aureus (STAPH-PLUS Pastorex, USA) included the following tests: Gram staining
and biochemical test of catalase, DNase, and the agglutination test.
Susceptibility
We have implemented the disk diffusion Kirby-Bauer method according to the Clinical
and Laboratory Standards Institute (CLSI) in order to identify the antimicrobial susceptibility.
We evaluated the susceptibility to oxacillin, ciprofloxacin, erythromycin, ampicillin-sulbactam,
clindamycin, and gentamicin as well. The susceptibility to oxacillin was tested through
the use of a cefoxitin disk considered as a breakpoint for resistance if the growing
inhibition zone of diameter ,21mm. Susceptibility to vancomycin was not evaluated
due to lack of access to methods to determine the minimum inhibitory concentration.
Intermediate susceptibility results were considered resistant.
Molecular Methods
We confirmed isolates as S. aureus molecularly through the amplification of a specific
16S rRNA gene, using the protocol and oligonucleotides described by Manfredi et al.
201011. We have extracted DNA from isolated colonies on blood agar by using the commercial
kit Wizard Genomic (Promega, USA) following the manufacturer instructions. DNA quantification
was performed using the UV spectrophotometer Biowave DNA (Cambridge, UK). The analysis
of the genetic variability of S. aureus isolates was carried out by the MLVA technique.
This technique involves amplifying seven loci using oligonucleotides and was previously
described by Sabat and collaborators.12 The strain ATCC®29213, negative mecA and producing weak-lactamase was included as
a control in each MLVA assay. We separated the PCR products on 7.5% polyacrylamide
gels and performed silver staining following the protocol described by Sambrook et
al.13 We captured digital images with the Kodak Digital Science team CD120 system (Kodak,
NYC, United States). We used TreeCon 1.3b software (Ghent University, Gante, Belgium)
for the analysis of the band patterns generated and the design of dendrograms.
Statistical issues
We have compared continuous variables such as: patients age, years undergoing hemodialysis,
white blood cell count, differential count of lymphocytes, neutrophils, and eosinophils
(expressed as number of cells/mm3) between patients with MRSA carriage versus patients
without MRSA carriage through the Student’s t-test; and dichotomous variables as well
such as: patients sex, day and turn of hemodialysis, use of antibiotics over the last
six months, the use of an invasive device, and history of rhinitis were analyzed using
the chi-square test. Statistical significance was defined as a p value of p<0.05 using
SPSS 15.0 software (IBM, NYC, United States) for statistical analysis.
Results
Among the 28 HD patients from the Nephrology Department of Hospital de Clínicas who
were part of this study, 50% (14/28) of them were carriers of S. aureus. Antibiotic
susceptibility testing identified 92.9% (13/14) of MRSA carriage. Isolates were also
resistant to other antibiotics, especially erythromycin and ampicillin-sulbactam,
both with 78.6% (11/14). 42.9 % (6/14) of the isolates were sensible to Gentamicin
(Table 1). Isolates that showed resistance to more than 4 families of antibiotics were considered
multiresistant and included 35.7% (5/14) of MRSA isolates (Table 2).
Table 1.
Clinical outcomes when comparing the interventions.
|
Sensible % (n) |
Resistance % (n) |
| Oxacillin |
7%(1) |
93%(13) |
| Erythromycin |
21%(3) |
79%(11) |
| Ciprofloxacin |
29%(4) |
71%(10) |
| Clindamycin |
36%(5) |
64%(9) |
| Gentamicin |
43%(6) |
57%(8) |
Table 2.
Resistance profile of Staphylococcus aureus isolates.
|
Percentage (n) |
| More than 1 antibiotic classes |
93%(13) |
| More than 2 antibiotic classes |
86%(12) |
| More than 3 antibiotic classes |
50% (7) |
| More than 4 antibiotic classes |
36% (5) |
The analysis of the demographic and clinical data of the hemodialyzed patients classified
as: S. aureus carriers and non-carriers showed that only the differences registered
in rhinitis OR=6.6 (95% CI: 1.2 – 34.4, p=0.022) were statistically significant (p<0.05)
(Table 3).
Table 3.
Demographics of Hemodialyzed Patients
|
Staphylococcus aureus Carriers (n=14) |
Staphylococcus aureus Non Carriers (n=14) |
| Age (Mean±SD)
|
41±14 |
44±18 |
| Dialysis Turn |
|
|
| Evening |
43% (6) |
71% (10) |
| Afternoon |
50% (7) |
29% (4) |
| Days of dialysis |
|
|
| Monday, Wednesday, Friday |
43% (6) |
71% (10) |
| Tuesday, Thursday, Saturday |
57% (8) |
29% (4) |
| Time of dialysis treatment in years (Mean±SD)
|
6.1±5.5 |
5.1±4.6 |
| Use of Arteriovenous fistula |
79% (11) |
64% (9) |
| History of different Vascular Access (Mean±SD)
|
2±2 |
3±2 |
| Antibiotic treatment in the last 6 months |
36%(5) |
29% (4) |
| Rhinitisa |
64% (9) |
21% (3) |
| Laboratory data (Mean±SD)
|
|
|
| Leucocytes |
7510±3767 |
6012±1531 |
| Neutrophils |
5443±3609 |
3788±1143 |
| Lymphocytes |
1509±333 |
1602±572 |
| Eosinophils |
263±302 |
243±333 |
| Albumin (g/dL) |
3.7±0.24 |
4.01±0.69 |
| Hemoglobin (g/dL) |
9.74±1.71 |
9.08±2.03 |
aDifference was statistically significant.
All the isolates identified as S.aureus by biochemical methods (n=14) were confirmed
by the detection of 16S rRNA gene by PCR amplification, with a 100% agreement between
phenotypic and genotypic methods. PCR amplification of the 7 loci included in the
MLVA method were optimal for cluster analysis in 7 of the 14 MRSA isolates in the
study and it gave us the chance to determine the genetic variability (Table 4). The bioinformatic analysis of the band profiles obtained in these isolates generated
a dendrogram which is showed in Figure 1. This graphic display has the form of a tree, in which the distance between braches
indicates the genetic difference. It can be observed that isolates identified as CR-4
and CR-5 show the same band profile and are clustered together in one branch at the
same distance, whereas isolates CR-10 and CR-12, and CR-9 and CR-14 as well, show
similar but not identical band profiles and are clustered in groups with branches
relatively close to each other (Figure 1).
Table 4.
Phenotypical and Genotypical Characteristics of Isolates Typed by MLVA.
| Patient |
Sex |
Age |
HD Turn |
Isolate |
O |
E |
A |
L |
G |
C |
MLVA Cluster |
| 4 |
M |
53 |
Afternoon |
CR-4 |
R |
R |
R |
R |
R |
R |
A |
| 5 |
M |
38 |
Afternoon |
CR-5 |
R |
R |
R |
R |
R |
R |
A |
| 9 |
F |
57 |
Evening |
CR-9 |
R |
R |
R |
I |
R |
I |
E |
| 10 |
M |
58 |
Afternoon |
CR-10 |
I |
I |
I |
R |
S |
S |
B |
| 12 |
M |
37 |
Afternoon |
CR-12 |
S |
S |
S |
S |
S |
S |
C |
| 14 |
F |
32 |
Evening |
CR-14 |
R |
S |
I |
S |
I |
I |
F |
| 17 |
M |
20 |
Evening |
CR-17 |
R |
R |
R |
R |
R |
R |
D |
MLVA: Multiple Locus Variable Number of Tandem Repeat Analysis; HD: Hemodialysis;
CR: Sample code; M=Male; F=Female; R=Resistant; I=Intermediate; S=Sensitive; O=Methicillin;
E=Erythromycin; A=Ampicillin-sulbactam; L=Clindamycin; G=Gentamicin; C=Ciprofloxacin.
Figure 1.
Genetic Relationship Observed by MLVA among Staphylococcus aureus Isolates
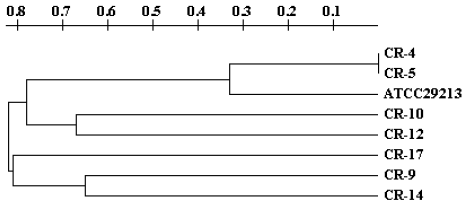
Legend: Dendrogram obtained with the software TreeCon v1.3b. Seven offourteen isolates
were processed by the MLVA technique for cluster analysis including a control ATCC®29213).
The dendrogram shows the relationship between isolates, the abscissa numbers are indicative
of the percentage of genetic variability. For example, between isolates CR-4 and CR-5
there is no variability, but between CR-4 and CR-17 there is 80% genetic variability.
Discussion
The results showed an alarming frequency of Staphylococcus aureus carriage in hemodialyzed
patients (50%), considering that carrying this organism is an important cause of morbidity
and mortality in patients who receive hemodialysis. This study portrayed a high prevalence
of nasal S. aureus carriage compared with others studies14,15 that showed between 5-13% of S. aureus carriage.14,15 However, two studies demonstrated similar carriage rates of S. aureus in hemodialysis
patients including Verhoeven et al (58%)16 and Price et al (49%).17 The risk of infections in nasal carriers of this microorganism is real and well defined.18 It is important to point out that the small number of patients included in this study
is a limitation and that the results cannot be extrapolated to other hemodialysis
services. However, the data regarding this topic is the first published in Paraguay
and further research or systematic studies could be done throughout the country in
the future.
There are several studies on MRSA carriage, which reported variable data. One meta-analysis
by Zacharioudakis et al.19 found 6% of carriage; the frequency in this study was much
higher (46.4%). MRSA carriers had an increased risk of mortality in all diseases.
This is attributable to the characteristics of the patients that are carriers of MRSA,
who exhibit an impaired immune response, and as a consequence lead to an increased
risk of infections.8 Risk factors predispose not only for MRSA carriage, but also for multidrug-resistant
gram negative bacilli (GNB) and vancomycin-resistant enterococci (VRE).20 There is the possibility of vancomycin resistance in MRSA, through the transmission
of the vanA gene, as occurred in neighboring countries.21 In accordance with the high level of MRSA carriage, 32% of the isolates in the study
were resistant to all antibiotics used in the Kirby-Bauer test, especially erythromycin
(79%). Gentamicin was the antibiotic which had the most effective action in-vitro
(43% sensitivity), but the resistance rates were high as well. This study showed similar
rates of resistance compared to other countries within the region.22 The high rates of resistance to antibiotics by MRSA display the capability of the
pathogen to carry more resistance genes than MSSA.
It is important to mention that we did not test the susceptibility to vancomycin in
this study; this would be relevant for the epidemiological surveillance and behavior
of strains in the hospital environment. In our country, VISA (vancomycin-intermediate
S. aureus) or hVISA (heterogeneous vancomycin-intermediate S. aureus) have not been
reported yet. This issue could be associated with the restricted access to an automated
method to test vancomycin resistance by the minimum inhibitory concentration (MIC)
method that is not available in all microbiology laboratories.
By using the MLVA technique, we identified 2 isolates that showed identical band profiles.
These isolates also shared the same antimicrobial spectrum (CR-4, CR-5), showing the
possibility that they correspond to the same clone. This should be analyzed by the
combination of other molecular methods such as field gel electrophoresis (PFGE), multilocus
sequencing typing and spa typing.
We discarded the possibility of epidemiological outbreaks due to the low number of
isolates having identical MLVA profiles, but we have not excluded any risk of transmission
between patients in the service. Internal transmission taking place in the hemodialysis
service is a risk, considering that multi-resistant bacteria are circulating. The
major goal of MLVA is to determine if isolates are epidemiologically linked and followed
over a relatively short period of time to see if they are related or unrelated. In
this study, MLVA was helpful for discarding outbreaks in a hospital setting. MLVA
is useful, fast, easy to perform, and particularly cheaper to the PFGE method already
used in PCR-based assays.23,24 In respect to other variables that were analyzed in patients, the highest frequency
of rhinitis may be related to the fundamental role of eosinophils in the production
of itching and predisposition to allergic reactions.25 Additionally, the nasal carriage of S. aureus produces an immunomodula-tory effect
that could contribute to airway inflammation and allergic response in patients with
allergic rhinitis.26 In the case of patients undergoing hemodialysis, presenting symptoms or signs may
be overlapped by uremic pruritus.27 Study limitations were the low number of samples included as it was difficult to
extrapolate results to all patients undergoing hemodialysis. Nevertheless, as a pilot
study this generated the first set of data on the characterization of S. aureus from
hemodialysis patients in Paraguay. As a perspective, we are interested in extending
the study to other centers throughout the country.
The spread of S. aureus can be controlled through reinforcement of appropriate use
of antibiotics, hand washing and laboratory surveillance for S. aureus, particularly
in the nosocomial wards, in order to identify sources of outbreaks. The application
of MLVA could help us to elucidate if isolates are epidemio-logically linked, useful
information for confirming or discarding outbreaks in the hospital environment.28,29
Acknowledgments
Special thanks to Dr. Marcos Martinez and the staff of the Nephrology Department of
Hospital de Clínicas (Asuncion, Paraguay) for their support at the interviews with
the patients and the kindly provided permission to access the clinical data.
Conflict of Interest Statement & Funding
None of the authors shows any conflicts of interest during the investigation or publication
of this article.
Author Contributions
Conception and design the work/idea: RASI, CRCV, GAVR, JP, RMGF. Collect data/obtaining
results: RASI, CRCV, GAVR. Analysis and interpretation of data: RASI, FR, RMGF. Write
the manuscript: RASI. Critical revision of the manuscript: FR, E. Approval of the
final version: RASI, FR, CRCV, GAVR, JP, RMGF. Contribution of patients or study material:
CRCV, GAVR, JP. Administrative or technical advice: FR, JP, RMGF..
References
1. Giarola LB, dos Santos RR, Tognim MCB, Borelli SD, Bedendo J. Carriage frequency, phenotypic and genotypic characteristics of Staphylococcus aureus
isolated from dialysis and kidney transplant patients at a hospital in Northern Paraná. Braz J Microbiol. 2012 Jul;43(3):923–30.
2. Greiner W, Rasch A, Köhler D, Salzberger B, Fätkenheuer G, Leidig M. Clinical outcome and costs of nosocomial and community-acquired Staphylococcus aureus
bloodstream infection in haemodialysis patients. Clin Microbiol Infect. 2007 Mar;13(3):264–8.
3. Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible
Clones of Staphylococcus aureus. J Clin Microbiol. 2000 Mar;38(3):1008–15.
4. Saxena AK, Panhotra BR. The Prevalence of Nasal Carriage of Staphylococcus aureus and Associated Vascular
Access-Related Septicemia Among Patients on Hemodialysis in Al-Hasa Region of Saudi
Arabia. Saudi J Kidney Dis Transpl. 2003 Jan;14(1):30–8.
5. Sanavi S, Ghods A, Afshar R. Catheter associated infections in hemodialysis patients. Saudi J Kidney Dis Transpl. 2007 Jan;18(1):43.
6. Vandecasteele SJ, Boelaert JR, De Vriese AS. Staphylococcus aureus Infections in Hemodialysis: What a Nephrologist Should Know. Clin J Am Soc Nephrol. 2009 Aug;4(8):1388–400.
7. Schmid H, Romanos A, Schiffl H, Lederer SR. Persistent nasal methicillin-resistant Staphylococcus aureus carriage in hemodialysis
outpatients: a predictor of worse outcome. BMC Nephrol. 2013 Apr;14:93.
8. Lai CF, Liao CH, Pai MF, Chu FY, Hsu SP, Chen HY, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus is associated with higher
all-cause mortality in hemodialysis patients. Clin J Am Soc Nephrol. 2011 Jan;6(1):167–74.
9. Peacock SJ, de Silva GD, Justice A, Cowland A, Moore CE, Winearls CG, et al. Comparison of Multilocus Sequence Typing and Pulsed-Field Gel Electrophoresis as Tools
for Typing Staphylococcus aureus Isolates in a Microepidemio-logical Setting. J Clin Microbiol. 2002 Oct;40(10):3764–70.
10. Chung S, Yi J, Jang MH, Joo S-I, Ra EK, Kim SY, et al. Comparison of Modified Multiple-locus Variable-number Tandem-repeat Fingerprinting
with Pulsed-field Gel Electrophoresis for Typing Clinical Isolates of Staphylococcus
aureus. Ann Lab Med. 2012 Jan;32(1):50–6.
11. Manfredi EA, Leotta GA, Rivas M. Multiplex PCR for the detection of sea, seb, sec, sed and see genes of Staphylococcus
aureus. Characterization of isolates from food. Rev Argent Microbiol. 2010 Sep;42(3):212–5.
12. Sabat A, Krzyszton-Russjan J, Strzalka W, Filipek R, Kosowska K, Hryniewicz W, et al. New Method for Typing Staphylococcus aureus Strains: Multiple-Locus Variable-Number
Tandem Repeat Analysis of Polymorphism and Genetic Relationships of Clinical Isolates. J Clin Microbiol. 2003 Apr;41(4):1801–4.
13. Sambrook J, Russell D. Molecular Cloning. A Laboratory Manual. 3rd ed. New York: Cold Spring Harbor Laboratory; 2001.
14. Jamil B, Bokhari MT, Saeed A, Mukhtar Bokhari MZ, Hussain Z, Khalid T, et al. Bacteremia: Prevalence and antimicrobial resistance profiling in chronic kidney diseases
and renal transplant patients. J Pak Med Assoc. 2016 Jun;66(6):705–9.
15. Mohajeri P, Azizkhani S, Farahani A, Norozi B. Genotyping of coa and aroA Genes of Methicillin-Resistant Staphylococcus aureus Strains
Isolated From Nasal Samples in Western Iran. Jundishapur J Microbiol. 2016 Jan;9(1):e26460.
16. Verhoeven PO, Gagnaire J, Haddar CH, Grattard F, Thibaudin D, Afiani A, et al. Identifying Hemodialysis Patients With the Highest Risk of Staphylococcus aureus Endogenous
Infection Through a Simple Nasal Sampling Algorithm. Medicine (Baltimore). 2016 Apr;95(14):e3231.
17. Price A, Sarween N, Gupta I, Baharani J. Meticillin-resistant Staphylococcus aureus and meticillin-susceptible Staphylococcus
aureus screening in a cohort of haemodialysis patients: carriage, demographics and
outcomes. J Hosp Infect. 2015 May;90(1):22–7.
18. Devraj A, Siva Tez Pinnamaneni V, Biswal M, Ramachandran R, Jha V. Extranasal Staphylococcus aureus colonization predisposes to bloodstream infections
in patients on hemodialysis with noncuffed internal jugular vein catheters. Hemodial Int. 2016 Jun;doi: http://dx.doi.org/10.1111/hdi.12450
 .
.
19. Zacharioudakis IM, Zervou FN, Ziakas PD, Mylonakis E. Meta-Analysis of Methicillin-Resistant Staphylococcus aureus Colonization and Risk
of Infection in Dialysis Patients. J Am Soc Nephrol. 2014 Sep; 25(9):2131–41.
20. Lim CJ, Cheng AC, Kennon J, Spelman D, Hale D, Melican G, et al. Prevalence of multidrug-resistant organisms and risk factors for carriage in long-term
care facilities: a nested case-control study. J Antimicrob Chemother. 2014 Jul;69(7):1972–80.
21. Rabelo MA, Bezerra Neto AM, Loibman SO, Lima JL, Ferreira EL, Leal NC, et al. The occurrence and dissemination of methicillin and vancomycin-resistant Staphylococcus
in samples from patients and health professionals of a university hospital in Recife,
State of Pernambuco, Brazil. Rev Soc Bras Med Trop. 2014 Aug;47(4):437–46.
22. Reyes J, Rincón S, Díaz L, Panesso D, Contreras GA, Zurita J, et al. Dissemination of Methicillin-Resistant Staphylococcus aureus USA300 Sequence Type
8 Lineage in Latin America. Clin Infect Dis. 2009 Dec;49(12):1861–7.
23. Tenover FC, Vaughn RR, McDougal LK, Fosheim GE, McGowan JE Jr. Multiple-Locus Variable-Number Tandem-Repeat Assay Analysis of Methicillin-Resistant
Staphylococcus aureus Strains. J Clin Microbiol. 2007 Jul;45(7):2215–9.
24. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, et al. Comparison of Multiple-Locus Variable-Number Tandem-Repeat Analysis with Pulsed-Field
Gel Electrophoresis, spa Typing, and Multilocus Sequence Typing for Clonal Characterization
of Staphylococcus aureus Isolates. J Clin Microbiol. 2005 Jul;43(7):3095–100.
25. Garnacho-Montero J, Huici-Moreno MJ, Gutiérrez-Pizarraya A, López I, Márquez-Vácaro JA, Macher H, et al. Prognostic and diagnostic value of eosinopenia, C-reactive protein, procalcitonin,
and circulating cell-free DNA in critically ill patients admitted with suspicion of
sepsis. Crit Care. 2014 Jun;18(3):R116.
26. Gröger M, Bernt A, Wolf M, Mack B, Pfrogner E, Becker S, et al. Eosinophils and mast cells: a comparison of nasal mucosa histology and cytology to
markers in nasal discharge in patients with chronic sino-nasal diseases. Eur Arch Otorhinolaryngol. 2013 Sep;270(10):2667–76.
27. Çevik C, Yula E, Yengil E, Gülmez Mβ, Akbay E. Identification of nasal bacterial flora profile and carriage rates of methicillin-resistant
Staphylococcus aureus in patients with allergic rhinitis. Eur Arch Otorhinolaryngol. 2014 Jan;271(1):103–7.
28. Mettang T, Kremer AE. Uremic pruritus. Kidney Int. 2015 Apr;87(4):685–91.
29. Kim YC, Kim MH, Song JE, Ahn JY, Oh DH, Kweon OM, et al. Trend of methicillin-resistant Staphylococcus aureus (MRSA) bacteremia in an institution
with a high rate of MRSA after the reinforcement of antibiotic stewardship and hand
hygiene. Am J Infect Control. 2013 May;41(5):e39–43.
Rubén Arturo Silvero-Isidre, 1 Universidad Nacional de Asunción, Paraguay.
Fátima Rodríguez-Acosta, 1 Universidad Nacional de Asunción, Paraguay.
César Rodrigo Cristaldo-Vargas, 1 Universidad Nacional de Asunción, Paraguay.
Genaro Américo Velázquez-Romero, 1 Universidad Nacional de Asunción, Paraguay.
José Félix Plans-Perrota, 1 Universidad Nacional de Asunción, Paraguay.
Rosa María Guillén-Fretes, 1 Universidad Nacional de Asunción, Paraguay.
Mihnea-Alexandru Găman, Editor
About the Author: Arturo Silvero Isidre is a final-year medical student of a six-year program at the
Universidad Nacional de Asunción, Paraguay. He is editor of a scientific journal called
CIMEL and a member of a scientific society of medical students (SOCIEM-UNA) in Paraguay.
Correspondence Rosa María Guillén Fretes, Email: rmguillenf@gmail.com
Cite as: Silvero Isidre RA, Rodríguez Acosta F, Cristaldo Vargas CR, Velázquez Romero GA, Plans Perrota JF, Guillén Fretes RM. Molecular Characterization of Staphylococcus aureus Isolates Obtained from Hemodialyzed Patients at the Hospital de Clínicas of Paraguay: A pilot study. Int J Med Students. 2017 Jan-Apr;5(1):1419.
Copyright © 2017 Rubén Arturo Silvero-Isidre, Fátima Rodríguez-Acosta, César Rodrigo
Cristaldo-Vargas, Genaro Américo Velázquez-Romero, José Félix Plans-Perrota, Rosa
María Guillén-Fretes
International Journal of Medical Students, VOLUME 5, NUMBER 1, April 2017

