Original Articles
Assessment of Antimicrobial Resistance and Susceptibility Pattern of UTI-causing Microorganisms
in Southern Punjab, Pakistan
Muhammad Mehwar Anjum1, Faiza Khalid1, Rida Saleem1, Muhammad Awais Bin Abdul Malik1, Muhammad Rizwan1
doi: http://dx.doi.org/ijms.2024.2163
Volume 12, Number 3: 267-273
Received 10 07 2024;
Rev-request 20 10 2023;
Rev-request 13 07 2024;
Rev-recd 11 03 2024;
Rev-recd 13 07 2024;
Accepted 14 07 2024
ABSTRACT
Background:
Bacterial resistance against antibiotics has become a global challenge and measures
are needed to stop this. The aim of this study is to highlight this problem and to
determine the antibiotic susceptibility pattern of organisms in Southern Punjab, Pakistan.
Method:
This descriptive cross-sectional study was conducted in Sheikh Zayed Medical Hospital,
Rahim Yar Khan. The urine samples obtained from 4 different wards were sent for culture
and sensitivity analysis. 9 antibiotics (Nitrofurantoin, Fosfomycin, Ciprofloxacin,
Ceftriaxone, Trimethoprim-Sulfamethoxazole, Norfloxacin, Linezolid, Amoxicillin, and
Imipenem) were tested against 5 isolated strains of uropathogens using Kirby Bauer
disk diffusion test. The sensitivity reports were obtained, and data points were entered
into a spreadsheet and analysed using SPSS.
Results:
Out of 101 samples of uropathogens that showed positive growths (42.08%), 53 (52.4%)
were from male patients and 48 (47.5%) positive growths were from females. Escherichia Coli had the highest positive growths (58%) followed by Pseudomonas (19%) Klebsiella (13%), Staphylococcus Aureus (7%) and Coagulase-negative staphylococci (3%). Imipenem was the most sensitive drug whereas the highest resistance by organisms
was developed against TMP-SMX. No significant association (p>0.05) was found between
any of the anti-microbial drugs and Escherichia coli, gram-positive uropathogens, and gram negative uropathogens.
Conclusion:
The high increasing rate of broad-spectrum antibiotics resistance suggests that diagnostic
and culture tests should be encouraged in hospitals. Based on these test results,
appropriate antibiotics should be prescribed. The limitations include the inability
to distinguish between nosocomial and community-acquired urinary tract infections
and also did not consider other demographic factors like age.
Introduction
Antibiotics play a vital role in the treatment of infectious diseases by bacterial
stasis or lysis. However, the increased and improper use of antibiotics has led to
the development of antibiotic resistance, where bacteria acquire the ability to survive
antibiotic treatment.1 Bacteria employ various mechanisms to develop resistance, including enzyme production,
reduced drug sensitivity, and possession of numerous mobilizable genes within bacterial
populations. Additionally, patient self-medication, over-prescribing, and incomplete
dosing significantly contribute to bacterial resistance.2
In the context of antibiotic resistance, Pakistan, as a developing nation, faces the
challenge of multiple drug-resistant and extensively drug-resistant bacteria.3 An example of this issue is the synergistic action of Azithromycin and fluoroquinolones
in treating co-infections alongside COVID-19, which has led to the associated overuse
of antibiotics and the potential for antimicrobial resistance.4 Given the prevalent misuse and overuse of antibiotics in Pakistan, understanding
susceptibility patterns and developing effective strategies are imperative, especially
since there has been no research of this kind conducted in the lower Punjab region,
necessitating immediate action to establish efficient measures.
Urinary tract infections (UTIs) are prevalent infectious diseases, particularly in
developing countries. The emergence of drug resistance among bacterial uropathogens
has further complicated the problem, giving rise to antibiotic-resistant species.5 The 2017–2018 GLASS (Global Antimicrobial Resistance and Use Surveillance System)
report indicated over 70% resistance to ceftriaxone and ciprofloxacin in Escherichia
coli in Pakistan. To tackle this issue effectively, it is crucial to determine the
susceptibility patterns of uropathogens to specific antibiotics to minimize exacerbation
of resistance. Moreover, the implementation of antibiotic stewardship programs is
essential to enhance healthcare quality and ensure appropriate antibiotic use.6 These collective measures will enable healthcare professionals to manage urinary
tract infections more effectively, employing a targeted approach that reduces recurrence
rates and achieves higher cure rates within shorter durations.
The primary objective of our study is to identify UTI causing pathogens and their
antimicrobial susceptibility patterns based on gender, gram-staining, and hospital
ward. This research aims to improve treatment efficacy and reduce recurrence rates.
Additionally, our study seeks to promote culture testing and the practice of appropriate
prescription of antibiotics based on culture and susceptibility patterns. By addressing
these objectives, we aim to contribute valuable insights that will aid in the development
of more effective treatment strategies for UTIs, while also improving appropriate
antibiotic prescribing practices.
Methods
Study Design and Setting
The study was a descriptive cross-sectional study conducted at Sheikh Zayed Medical
Hospital in Rahim Yar Khan, Punjab, Pakistan from May to July 2022. The purpose of
the study was to investigate the prevalence of urinary tract infections (UTIs) in
patients admitted to the medicine, gynecology, surgery, and nephrology wards. These
departments were chosen due to the high burden of UTI patients in them, as observed
by the researchers during their hospital rotations. The study was evaluated using
the STROBE checklist for cross sectional studies. Ethical clearance was obtained from
the Institutional Research Board Sheikh Zayed Medical College/Hospital, with the reference
No: 479/IRB/SZMC/SZH, and written permission was obtained from the head of each department.
The researchers had no potential conflicts of interest, and no external funding for
the study.
Participants
Patients presenting with uncomplicated UTI symptoms (abdominal pain, burning micturition,
cloudy or foul-smelling urine) were included in the study, while those at high risk
of complications or in critical condition were excluded. Immunocompromised, septic,
and patients with other comorbidities like diabetes and those who had taken antibiotics
in the last 24 hours were also excluded. This was done as such patients may require
different management approaches that may confound the results. This exclusion criterion
resulted in a higher proportion of female patients in the study, as the gynecology
ward primarily admitted female patients. Simple random sampling was employed.
Data Collection
Patients included in the study provided informed consent and their data was collected
using a self-developed questionnaire which included variables such as name, age, gender,
and ward name. The questionnaire was pre-tested through trial interviews to improve
question-asking methods and variables. Based on previous studies and hospital records,
a study sample size of more than 200 was expected between the period of study from
May to July 2022.
Confirmation of Diagnosis
Early morning mid-stream urine samples were collected from the patients and stored
in sterile urine collection containers. The urine culture samples were sent to the
microbiological culture sensitivity laboratory for analysis. Positive results were
determined when significant bacterial growth > 10^5 CFU/ml was observed. Colony study
and biochemical tests were performed to identify the microorganisms. Some of the disks
didn't show positive bacterial growth, which could be due to other microorganisms
like fungi, however only UTI causing bacteria were studied in this study.
Antibiotic Sensitivity Testing
MacConkey agar was used to subculture the colonies and obtain pure growth of the microorganisms.
The Kirby Bauer disk diffusion test was conducted to assess the sensitivity of the
isolates to ten different antibiotics. The ten antibiotics used in the procedure were
furnished separately as discs to the laboratory by the researchers, exclusively for
use on these urine samples. The measurement of the zone of inhibition of bacterial
growth was performed, and the results were compared with the guidelines of the Clinical
and Laboratory Standards Institute (CLSI). All intermediate results were considered
as sensitive too.
Antibiotics and Sensitivity Reports
The organisms were subjected to various groups of antibiotic discs from Oxoid, including
Nitrofurantoin (300µg), Fosfomycin (50µg), Ciprofloxacin (5µg), Ceftriaxone (30µg),
Trimethoprim/Sulfamethoxazole (1:19 and 25µg), Norfloxacin (5µg), Linezolid (30µg),
Amoxicillin (30µg), and Imipenem (10µg). The sensitivity reports of all patients were
individually studied. Then the data points were entered into a spreadsheet.
Statistical Analysis
The data was analyzed using IBM's Statistical Package for Social Sciences (SPSS version
26). Descriptive statistics, including numbers, frequencies, and percentages, were
used to describe the data. There was no missing data. Age was the only quantitative
variable. Tables and charts were utilized to display the data, and the association
between categorical variables was assessed using the Chi-square test. A standard p-value
of <0.05 was considered statistically significant. The chi-square test was also conducted
to assess the significance of antimicrobial drugs against gram-positive cocci (GPC)
and gram-negative bacteria separately. No sensitivity analysis was applicable.
Results
The study analyzed 240 urine samples from suspected urinary tract infection cases
admitted to the departments of medicine, gynecology, surgery, and nephrology. There
was no attrition and no missing data or loss of patients to follow-up between admission
and diagnosis and arrival of antimicrobial culture and sensitivity report. Among the
101 positive growths for uropathogens, 53 were male patients, and 48 were female patients.
The most common organism was Escherichia Coli (E. coli) with a growth rate of 58%, followed by Pseudomonas (19%), Klebsiella (13%), Staphylococcus Aureus (S. Aureus) (7%), and Coagulase-negative staphylococci (CoNS) (3%). Table 1 presents the growth rate of uropathogens and the drugs to which they are sensitive.
Figure 1 illustrates the distribution of microorganisms by gender of patients. The medicine
ward had the highest growth rate (45%), followed by gynecology (41.86%), surgery (41.17%),
and nephrology (38.75%). The uropathogens were further tested for antimicrobial susceptibility
patterns. The results of urine culture analysis from the samples collected in these
wards are summarized in Figure 2, presenting the growth rates and distribution of these organisms.
Table 1.
Microorganisms and their Sensitivity in Percentage to Various Antibiotics.
| Microorganism |
Total Growth |
Antibiotic Used |
| NFa |
CIPb |
FOSc |
CROd |
SXTe |
NORf |
LNZg |
AMCh |
IMPi |
| Escherichia Coli |
59 |
45.76 |
42.37 |
50.84 |
55.93 |
27.11 |
37.28 |
45.76 |
50.84 |
77.96 |
| Pseudomonas |
19 |
36.84 |
31.57 |
42.10 |
42.10 |
21.05 |
26.31 |
63.15 |
31.57 |
68.42 |
| Klebsiella |
13 |
38.46 |
30.76 |
30.76 |
46.15 |
30.76 |
38.46 |
53.8 |
46.15 |
76.92 |
| S. Aureus |
7 |
42.85 |
28.57 |
57.14 |
57.14 |
28.57 |
28.57 |
42.85 |
57.14 |
71.42 |
| CoNSj |
3 |
33.33 |
33.33 |
66.66 |
66.66 |
0 |
33.33 |
33.33 |
66.66 |
100 |
|
101 |
|
|
|
|
|
|
|
|
|
Legend:
aNitrofurantoin,
bCiprofloxacin,
cFosfomycin,
dCeftriaxone,
eTrimethoprim-Sulfamethoxazole,
fNorfloxacin,
gLinezolid,
hAmoxicillin,
iImipenem
jCoagulase-negative staphylococci (CoNS).
The numbers indicate in percentage the sensitivity of that uropathogen to the antibiotic
used. For instance, in E. Coli 45.76% percentage of the positive growth cultures were
sensitive to Nitrofurantoin and so on.
Figure 1.
Distribution of the Grown Microorganisms by Sex.
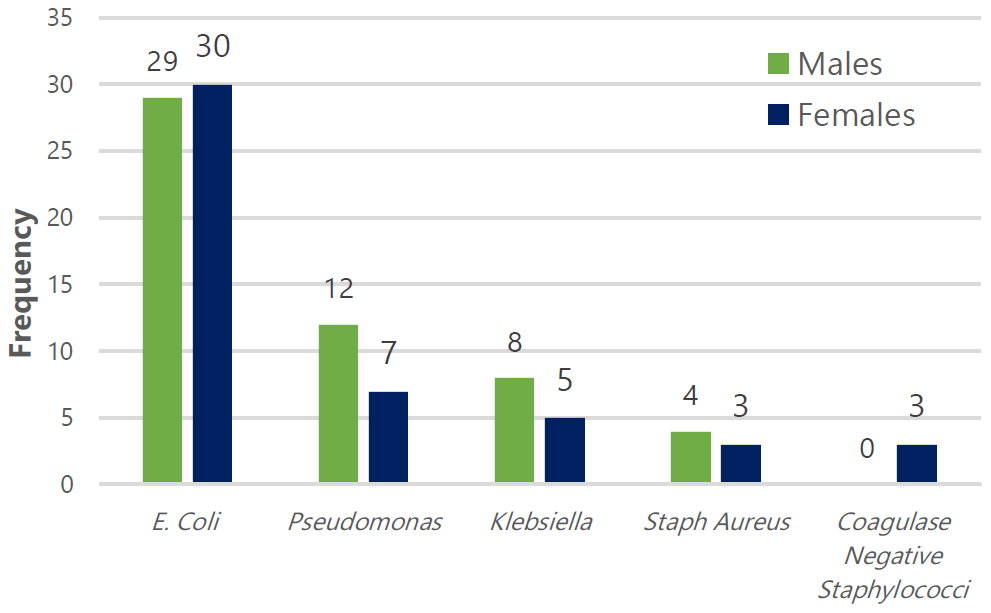
Legend: The distribution of microorganisms by patient sex includes 59 patients with Escherichia coli (29 males, 30 females), 19 with Pseudomonas (12 males, 7 females), 13 with Klebsiella (8 males, 5 females), 7 with Staphylococcus aureus (4 males, 3 females), and 3 with coagulase-negative staphylococci (0 males, 3 females).
Figure 2.
Growth Results by Ward of Sample Origin.
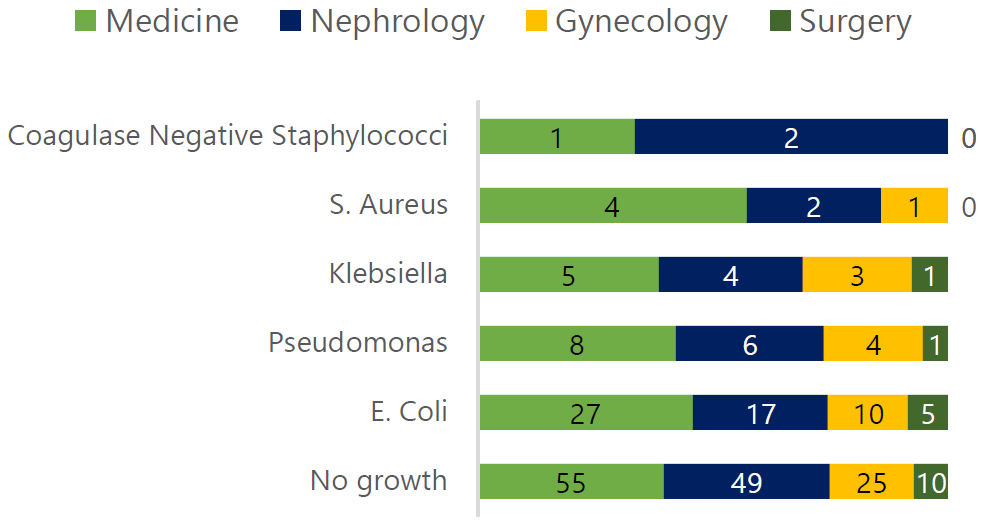
Legend: Uropathogens isolated from urine cultures by ward: Medicine ward had 45 positive
growths—27 E. coli, 8 Pseudomonas, 5 Klebsiella, 4 Staphylococcus aureus, and 1 coagulase-negative staphylococci. There were no coagulase-negative staphylococci
in the Gynecology or Surgery wards, and no Staphylococcus aureus causing UTIs in the Surgery ward.
In this study, a total of 9 antibiotics, including Nitrofurantoin (NF), Ciprofloxacin
(CIP), Fosfomycin (FOS), Ceftriaxone (CRO), Trimethoprim-Sulfamethoxazole (SXT), Norfloxacin
(NOR), Linezolid (LNZ), Amoxicillin (AMC), and Imipenem (IMP), were checked for their
sensitivity and resistance against the 5 organisms: E. coli, Pseudomonas, Klebsiella, S. Aureus, and CoNS.
Among gram-negative organisms, Imipenem showed the highest sensitivity (76%), while
the highest resistance was observed against TMP-SMX (74%). The findings of resistance
and sensitivity to these antibiotics by all uropathogens in general are summarized
in Figure 3. Gram-positive cocci demonstrated the highest sensitivity to Imipenem (80%) and the
highest resistance to TMP-SMX (80%). Their sensitivity and resistance to other antibiotics
is depicted in Figure 4. Notably, there was a difference in sensitivity between gram-positive cocci and gram-negative
uropathogens, particularly in relation to linezolid, as well as quantitative differences
among Amoxicillin, Fosfomycin, and Ceftriaxone.
Figure 3.
Effectiveness of Various Antimicrobial Drugs Against Common Gram-Negative Pathogens.
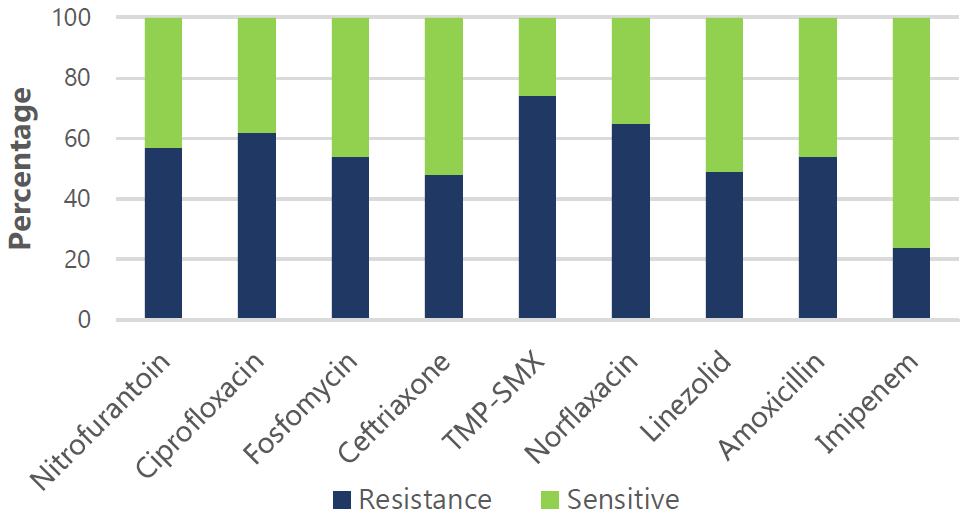
Legend: Resistance (R) and sensitivity (S) rates: Nitrofurantoin (57% R, 43% S), Ciprofloxacin
(62% R, 38% S), Fosfomycin (54% R, 46% S), Ceftriaxone (48% R, 52% S), TMP-SMX (74%
R, 26% S), Norfloxacin (65% R, 35% S), Linezolid (49% R, 51% S), Amoxicillin (54%
R, 46% S), and Imipenem (24% R, 76% S).
Figure 4.
Effectiveness of Various Antimicrobial Drugs Against Common Gram Positive Uropathogens.
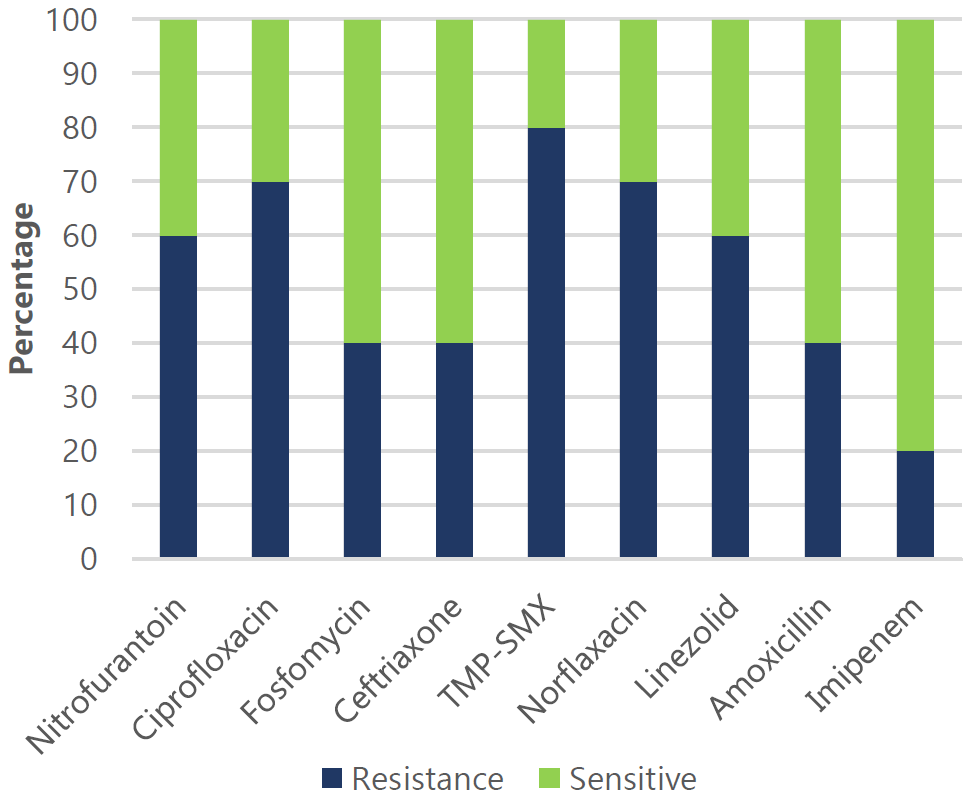
Legend: Resistance (R) and sensitivity (S) rates: Nitrofurantoin (60% R, 40% S), Ciprofloxacin
(70% R, 30% S), Fosfomycin (40% R, 60% S), Ceftriaxone (40% R, 60% S), TMP-SMX (80%
R, 20% S), Norfloxacin (70% R, 30% S), Linezolid (60% R, 40% S), Amoxicillin (40%
R, 60% S), and Imipenem (20% R, 80% S).
Regarding specific organisms, E. coli exhibited the highest sensitivity to IMP (78%) and the highest resistance against
SXT (73%). Pseudomonas displayed the highest resistance against SXT (79%), while it showed high sensitivity
to both LNZ (63%) and IMP (68%). S. Aureus isolates exhibited the highest resistance against CIP, SXT, and NOR (71%), while
its sensitivity was highest against IMP (71%), followed by FOS and CRO (50%). Klebsiella showed the highest sensitivity to IMP (76.92%), followed by LNZ (53.8%), CRO, AMC
(46.15%), and NF (38.46%). It displayed the highest resistance against SXT, CIP, and
FOS (69.24%).
CoNS showed maximum sensitivity to IMP (100%) and maximum resistance to SXT (100%) among
the few samples that showed its growth. The antimicrobial sensitivity pattern of E. coli based on the tissue culture sensitivity report is depicted in Figure 5, while Figure 6 presents the antimicrobial sensitivity pattern for Pseudomonas, and Figure 7 reports the susceptibility pattern of S. aureus.
Figure 5.
EAntimicrobial Drug Sensitivity and Resistance Pattern for E. Coli
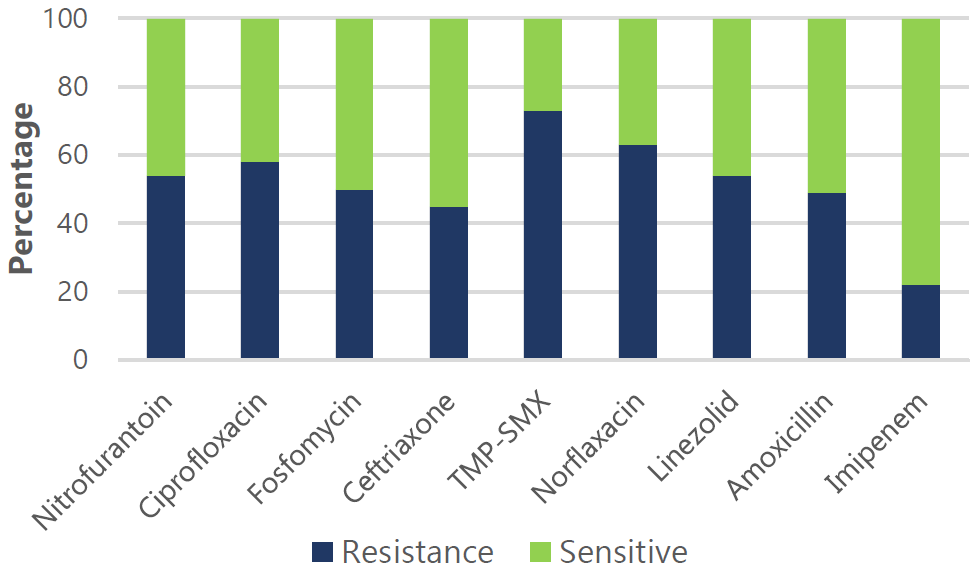
Legend: Resistance (R) and sensitivity (S) rates: Nitrofurantoin (54% R, 46% S), Ciprofloxacin
(58% R, 42% S), Fosfomycin (50% R, 50% S), Ceftriaxone (45% R, 55% S), TMP-SMX (73%
R, 27% S), Norfloxacin (63% R, 37% S), Linezolid (54% R, 46% S), Amoxicillin (49%
R, 51% S), and Imipenem (22% R, 78% S).
Figure 6.
Antimicrobial Drug Sensitivity and Resistance Pattern for Pseudomonas.
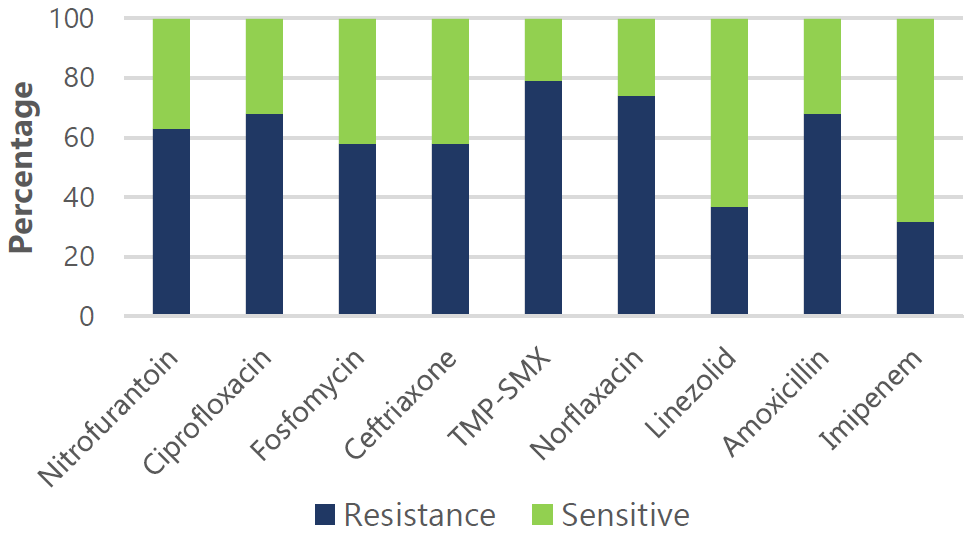
Legend Resistance (R) and sensitivity (S) rates: Nitrofurantoin (63% R, 37% S), Ciprofloxacin
(68% R, 32% S), Fosfomycin (58% R, 42% S), Ceftriaxone (58% R, 42% S), TMP-SMX (79%
R, 21% S), Norfloxacin (74% R, 26% S), Linezolid (37% R, 63% S), Amoxicillin (68%
R, 32% S), and Imipenem (32% R, 68% S).
Figure 7.
Antimicrobial Drug Sensitivity and Resistance Pattern for S. Aureus.
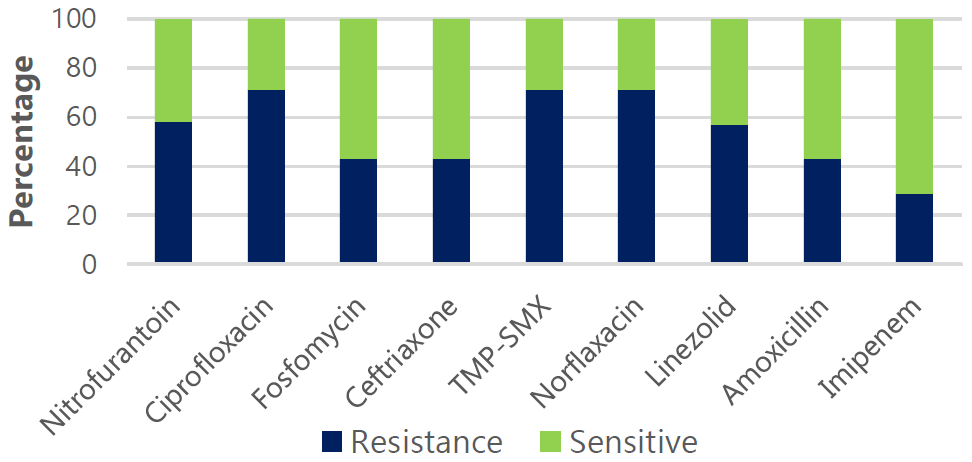
Legend Resistance (R) and sensitivity (S) rates: Nitrofurantoin (58% R, 42% S), Ciprofloxacin
(71% R, 29% S), Fosfomycin (43% R, 57% S), Ceftriaxone (43% R, 57% S), TMP-SMX (71%
R, 29% S), Norfloxacin (71% R, 29% S), Linezolid (57% R, 43% S), Amoxicillin (43%
R, 57% S), and Imipenem (29% R, 71% S).
With the Chi-square test, no significant association was found between any of the
antimicrobial drugs and E. coli growth or sensitivity to Imipenem. No statistically significant findings were also
observed in the efficacy of antimicrobial drugs against Gram-positive cocci (GPC)
and Gram-negative bacteria separately.
Discussion
In the present study, we observed an increasing trend of resistance to commonly used
antibiotics among uropathogens. The culture of bacteria revealed a positivity ratio
of approximately 42%, with multiple isolated strains exhibiting resistance to the
tested drugs. The susceptibility pattern differed between gram-positive and gram-negative
organisms. Gram-negative organisms showed increasing resistance to traditional urinary
tract infection antibiotics such as trimethoprim-sulfamethoxazole, norfloxacin, and
ciprofloxacin, while most of them remained susceptible to ceftriaxone, linezolid,
and imipenem. Gram-positive organisms demonstrated moderate susceptibility to ceftriaxone,
fosfomycin, and amoxicillin, with imipenem being the most effective. Fluoroquinolones
and other antibiotics had reduced effectiveness against most gram-positive strains.
Coagulase-negative staphylococci were only found in women, and the medicine ward exhibited
a higher incidence of urinary tract infections compared to other wards. E. coli affected more females than males, indicating a higher risk for females.7 Amoxicillin and fosfomycin showed relatively better efficacy against E. coli compared to other gram-negative bacteria, while linezolid was less effective. These
findings are consistent with previous research conducted in Europe.8 Pseudomonas displayed high resistance to all antibiotics except imipenem and linezolid. The resistance
observed can be attributed to various factors, including self-medication, overprescription,
insufficient doses, and inappropriate antibiotic usage.
Limitations of this study should be acknowledged. Firstly, the study was unable to
differentiate between nosocomial (hospital-acquired) urinary tract infections and
those caused through other colonization routes. As the urine cultures were obtained
from hospital patients, the identified microbes could represent multi-drug resistant
strains present in the hospital environment, acquired by the patients due to stress,
immunocompromise states, or other hospital-related factors.9 This could give rise to selection bias as the selected population was from hospital
settings and did not include patients treated in community settings. Additionally,
the male-to-female patient ratio is not truly representative due to some data being
primarily collected from the gynecology ward, which predominantly caters to women
patients. The study also did not consider factors such as age, patient awareness,
and habits related to self-medication, immune status, socio-economic status, hospital
stay and other co morbidities. These variables can act as confounding factors and
may create a bias. Human errors during antimicrobial disc sensitivity testing are
also possible. Although the chi-square test showed no significance between the usage
of any drug and the bacteria's sensitivity or resistance, further analysis with a
larger sample size is warranted to confirm these findings. Additional studies should
also be undertaken of patients with UTI with comorbidities, to study the disease trends
in them.
Although the study was well-designed and conducted with ethical considerations, there
is a chance of error in the procedures used, such as the disc sensitivity test and
the delay between disc application and incubation while measuring the reading zone,
creating a risk of measurement bias. This could be reduced by proper placing of the
disk and timely measurements of the reading zone.
The growth positivity rate observed in the study is within a reasonable range, indicating
the proficiency of the antimicrobial laboratory technique with minimal chances of
missing potential organism growth in the culture. E. coli and other gram-negative bacteria are commonly associated with UTIs due to their virulence
factors that facilitate colonization and ascending infection in the urinary tract.
E. coli, in particular, possesses specific Type I fimbriae and P pili containing hemolysin
and other toxins, contributing to its pathogenicity in causing urinary tract infections.
On the other hand, fewer gram-positive organisms were isolated in urine cultures of
UTIs. It is important to note that UTIs can occur in patients within hospitals, even
if they were not part of the initial presenting complaint.10 For instance, post-operative patients in surgery or gynecology wards, particularly
those who underwent caesarean section, were screened for urine culture, and some were
found to have infections.
The reduced resistance to Imipenem across all bacteria indicates appropriate prescription
practices for this drug and suggests a lower prevalence of carbapenemase-producing
bacteria, especially among E. coli and Klebsiella, in the region. However, this also suggests a higher occurrence of extended-spectrum
beta-lactamase (ESBL)-producing bacteria, rendering commonly used antibiotics for
other infectious diseases less effective. Previous in-vitro studies have also shown
increased effectiveness of Imipenem,11 but it is worth noting that research from India indicates resistance to multiple
carbapenems.12 It is important to reconsider the use of Trimethoprim-sulfamethoxazole (TMP-SMX)
as an antibiotic choice due to the high rate of resistance against it, despite its
supposed activity against Methicillin Resistant Staphylococcus Aureus (MRSA) and gram-negative bacteria causing UTIs. Fluoroquinolones, especially the
newer generations, were previously considered first-line therapy for complicated UTIs
and pyelonephritis. However, a considerable amount of literature now refutes their
use in UTIs for various reasons.13 This overprescription of fluoroquinolones may have led to the development of resistance
not only against these drugs but also against nitrofurantoin, which was commonly used
in cystitis. Commonly used drugs like ceftriaxone and amoxicillin show average rates
of sensitivity and resistance, indicating that these drugs could become ineffective
if they continue to be self-medicated and prescribed without appropriate history and
microbial culture. Fosfomycin, on the other hand, still retains some effectiveness
against gram-positive strains, possibly due to its lesser usage and lower prevalence.14 The increasing resistance to linezolid may be attributed to the high prevalence of
MRSAand the drug being the first-line treatment for multi-drug resistant organisms,
leading to its overuse.
The findings from this study have significant implications and can be generalized,
particularly among hospital populations worldwide. By looking at the susceptibility
pattern of UTI causing pathogens various hospitals in the region should monitor their
prescribed antibiotics and reassess their usage. A need for more culture and sensitivity-based
practice would also be felt to reinforce the better prescription of antibiotics. Such
practices will be particularly useful in low-income areas or in populations struggling
with hygiene, both of which will have significant overlap with the area where the
study was conducted as well. E. coli, Klebsiella, and Pseudomonas, which were studied for their antimicrobial susceptibility patterns in urine cultures,
are causative agents of both community- and hospital-acquired infections.15 The increasing resistance to most antibiotics observed in these pathogens not only
affects patients with UTIs but also those with other infectious diseases such as pneumonia,
diarrhea, and sepsis, who may be prescribed commonly used antibiotics like ceftriaxone,
amoxicillin, and linezolid without experiencing improvement due to resistance against
them. To further explore these correlations and trends, future studies can investigate
the antimicrobial susceptibility patterns of other frequently encountered infectious
diseases. It would be valuable to isolate and identify specific strains of microorganisms
with these resistance patterns and conduct genomic studies to explore the underlying
mechanisms of resistance. Antimicrobial stewardship programs in hospitals should be
promoted and implemented, along with systematic reviews of antibiotic prescriptions
across all hospital wards.16 It is crucial to increase awareness among healthcare professionals regarding the
importance of microbial culture and sensitivity evaluation and to educate the general
population about the risks of self-medication and microbial resistance.17 Additionally, there is a need for the development of improved diagnostic features
and testing methods that can differentiate not only between bacterial and viral infections
but also among different bacterial infections. Global collaboration is essential to
establish effective management plans based on accurate diagnosis.
Summary – Accelerating Translation
Title: Assessment of Antimicrobial Resistance and Susceptibility Pattern of UTI-causing
microorganisms in Southern Punjab, Pakistan.
Main Problem to Solve: As use of antibiotics becomes commonplace without complete adherence or protocol,
bacteria have started to emerge that are resistant to traditionally prescribed antibiotics.
This study determines how some urinary tract infection causing organisms have become
resistant to some common antibiotics.
Aim of Study: The aim of the study is to enhance treatment approaches, improve prescription practices,
encourage microorganism culture and antibiotic sensitivity testing in hospital settings
as well as encouraging antibiotic stewardships. Our goal is to provide valuable statistics
regarding resistance patterns of UTI-causing microorganisms in the region as well
as raise awareness and encourage health professionals to exercise caution in their
prescription decisions and self-prescription among patients.
Methodology: Early morning mid-stream urine samples were collected from different wards of Sheikh
Zayed Hospital, Rahim yar Khan. Informed consent and data like age, and ward name
was collected through a self-developed questionnaire. Among those with positive growths
were sent to microbiological culture sensitivity laboratory for analysis against 5
common antibiotics: Nitrofurantoin, Ciprofloxacin, Norfloxacin, TMP-SMX, Imipenem,
Amoxicillin, Ceftriaxone, Fosfomycin, Linezolid. Kirby Bauer disk diffusion test was
conducted to assess the sensitivity of the isolates. Data was analyzed using SPSS
and the association between categorical variables was assessed using the Chi-square
test. Descriptive statistics, including numbers, frequencies, and percentages, were
used to describe the results obtained from culture and sensitivity tests.
Results: Out of the 101 urine samples tested, 42.08% showed positive growth of bacteria causing
urinary tract infections. The most common bacteria found was Escherichia coli (E. coli), which accounted for 58% of the positive growths followed by Pseudomonas (19%) and Klebsiella (13%), Staphylococcus aureus (S. aureus) (7%), and coagulase-negative staphylococci (CoNS) (3%)
Imipenem was found to be the most effective drug, with the highest sensitivity against
bacteria causing urinary tract infections. It showed a sensitivity rate of 80% against
gram-positive uropathogens, 78% against E. coli, and 76% against gram negative uropathogens. On the other hand, the bacteria showed
the highest resistance to TMP-SMX, with resistance rates of 80% in gram-positive uropathogens,
74% in gram-negative uropathogens, and 73% in E. coli.
No significant association of sensitivity or resistance was found between any of the
antimicrobial drugs and E. Coli or gram-positive uropathogens or gram negative uropathogens
Conclusion: The study concluded that urinary tract organisms displayed escalating resistance
to commonly used antibiotics and antimicrobial stewardship programs are needed in
hospitals along with development of improved diagnostic features and testing methods
to differentiate among bacterial infections and eventually prescription of antibiotics
accordingly.
Acknowledgments
Dr. Muhammad Shahbaz Hussain, Head of Department pathology, for his guidance and supervision.
Dr. Muhammad Sajjad, In-charge antimicrobial culture and sensitivity report lab, for
his assistance and knowledge
Conflict of Interest Statement & Funding
The Authors have no funding, financial relationships or conflicts of interest to disclose.
Author Contributions
Conceptualization: MMA, RS, and MABAM. Methodology: MMA, RS, MABAM, and MR. Software:
MMA. Validation: FK, RS, MABAM, and MR. Formal Analysis: MMA, and FK. Data Curation:
MMA, FK, RS. MABAM. and MR. Investigation: MMA, FK, MABAM, and MR. Resources: MMA,
FK, RS, and MABAM. Writing – Original Draft: MMA, FK, and MR. Writing – Review & Editing:
MMA, FK, RS, MABAM, and MR. Visualization: MMA, FK, RS, and MABAM. Project Administration:
MMA, FK, and RS. Funding Acquisition: MMA, FK, RS, MABAM, and MR.
References
1. Sulis G, Adam P, Nafade V, Gore G, Daniels B, Daftary A, et al Antibiotic prescription practices in primary care in low- and middle-income countries:
A systematic review and meta-analysis. PLoS Med. 2020;17(6):e1003139.
2. Abdelrazik E, El-Hadidi M. Tracking Antibiotic Resistance from the Environment to Human Health. Methods Mol Biol. 2023;2649:289–301.
3. Bilal H, Khan MN, Rehman T, Hameed MF, Yang X. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect Dis. 2021;21(1):244.
4. Ul Mustafa Z, Salman M, Aldeyab M, Kow CS, Hasan SS. Antimicrobial consumption among hospitalized patients with COVID-19 in Pakistan. SN Compr Clin Med. 2021;3(8):1691–5.
5. Muzammil M, Adnan M, Sikandar SM, Waheed MU, Javed N, Ur Rehman MF. Study of culture and sensitivity patterns of urinary tract infections in patients
presenting with urinary symptoms in a tertiary care hospital. Cureus. 2020;12(2):e7013.
6. Alemkere G, Tenna A, Engidawork E. Antibiotic use practice and predictors of hospital outcome among patients with systemic
bacterial infection: Identifying targets for antibiotic and health care resource stewardship. PLoS One. 2019;14(2):e0212661.
7. Tamadonfar KO, Omattage NS, Spaulding CN, Hultgren SJ. Reaching the end of the line: Urinary tract infections. Microbiol Spectr. 2019;7(3).
8. Kot B. Antibiotic Resistance Among Uropathogenic Escherichia coli. Pol J Microbiol. 2019;68(4):403–15.
9. Sante L, Lecuona M, Jaime-Aguirre A, Arias A. Risk factors to secondary nosocomial bacteremia to UTI in a tertiary hospital. Rev Esp Quimioter. 2019;32(4):311–6.
10. Kranz J, Schmidt S, Wagenlehner Florian and Schneidewind L. Catheter-associated urinary tract infections in adult patients. Dtsch Arztebl Int. 2020;117(6):83–8.
11. Karlowsky JA, Lob SH, Kazmierczak KM, Young K, Motyl MR, Sahm DF. In vitro activity of imipenem/relebactam against Enterobacteriaceae and Pseudomonas
aeruginosa isolated from intraabdominal and urinary tract infection samples: SMART
Surveillance United States 2015–2017. J Glob Antimicrob Resist. 2020;21:223–8.
12. Ravishankar U, P S, Thayanidhi P. Antimicrobial resistance among uropathogens: Surveillance report from south India. Cureus. 2021;13(1):e12913.
13. Chao YS, Farrah K. Fluoroquinolones for the Treatment of Urinary Tract Infection: A Review of Clinical
Effectiveness, Cost-Effectiveness, and Guidelines. Ottawa (ON): Canadian Agency for Drugs and Technologies in Health; 2019.
14. Ten Doesschate T, Kuiper S, van Nieuwkoop Cees and Hassing RJ, Ketels T, van Mens Suzan P and van den Bijllaardt W, van der Bij AK, et al Fosfomycin vs ciprofloxacin as oral step-down treatment for Escherichia coli febrile
urinary tract infections in women: A randomized, placebo-controlled, double-blind,
multicenter trial. Clin Infect Dis. 2022;75(2):221–9.
15. Miyoshi-Akiyama T, Tada T, Ohmagari N, Viet Hung N, Tharavichitkul P, Pokhrel BM, et al Emergence and Spread of Epidemic Multidrug-Resistant Pseudomonas aeruginosa. Genome Biol Evol. 2017;9(12):3238–45.
16. Majumder MAA, Rahman S, Cohall Damian and Bharatha A, Singh K, Haque M, Gittens-St Hilaire M. Antimicrobial stewardship: Fighting antimicrobial resistance and protecting global
public health. Infect Drug Resist. 2020;13:4713–38.
17. Redfern J, Bowater L, Coulthwaite L, Verran J. Raising awareness of antimicrobial resistance among the general public in the UK:
the role of public engagement activities. JAC Antimicrob Resist. 2020;2(1):dlaa012.
Muhammad Mehwar Anjum, 1 Fifth Year MBBS Medical Student. Sheikh Zayed Medical College and Hospital, Rahim
Yar Khan, Pakistan.
Faiza Khalid, 1 Fifth Year MBBS Medical Student. Sheikh Zayed Medical College and Hospital, Rahim
Yar Khan, Pakistan.
Rida Saleem, 1 Fifth Year MBBS Medical Student. Sheikh Zayed Medical College and Hospital, Rahim
Yar Khan, Pakistan.
Muhammad Awais Bin Abdul Malik, 1 Fifth Year MBBS Medical Student. Sheikh Zayed Medical College and Hospital, Rahim
Yar Khan, Pakistan.
Muhammad Rizwan, 1 Fifth Year MBBS Medical Student. Sheikh Zayed Medical College and Hospital, Rahim
Yar Khan, Pakistan.
About the Author: Include 1 Muhammad Mehwar Anjum is currently a final year MBBS medical student of
Sheikh Zayed Medical College, Rahim Yar Khan, Pakistan of a total of a five-year program.
He is also a recipient of a distinction award (>85% score) in Biochemistry, Pharmacology
and Special Pathology.
Correspondence: Muhammad Mehwar Anjum. Address: C899+333, Rahim Yar Khan, Punjab 64200, Pakistán.
Email: mehwar9anjum9@gmail.com
Editor: Francisco J. Bonilla-Escobar;
Student Editors: Sebastian Diebel, Aurele Berjo Takoutsing Dongmo & Rebecca Murerwa;
Proofreader: Amy Phelan;
Layout Editor: Julian A. Zapata-Rios;
Process: Peer-reviewed
Cite as
Anjum MM, Khalid F, Saleem R, Abdul Malik MAB, Rizwan M. Assessment of Antimicrobial
Resistance and Susceptibility Pattern of UTI-causing microorganisms in Southern Punjab,
Pakistan. Int J Med Stud. 2024 Jul-Sep;12(3):267-273.
Copyright © 2024 Jenna Kupa, Ren Bruguera, Nadia Agnoli, Alicia Agnoli, Liliana Melgoza,
Anna Portnoy, P. Suzanne Portnoy
This work is licensed under a Creative Commons Attribution 4.0 International License.
International Journal of Medical Students, VOLUME 12, NUMBER 3, July 2024






