Experience
New Frontiers in Biomedical Research: A Medical Student Perspective
Ben Sayer1
doi: http://dx.doi.org/10.5195/ijms.2018.311
Volume 6, Number 3: 120-122
Received 14 09 2018:
Accepted 01 10 2018
The Experience
When I was offered a placement in Professor Haniffa’s world-leading immunology laboratory
at Newcastle University, it was a one-time opportunity to gain an insight into a field
that is rapidly developing. The focus of her group is mononuclear phagocytes, a family
of immune cells including dendritic cells, monocytes and macrophages, in different
contexts. A degree in Biomedical Sciences at St George’s University of London had
given me lab experience using techniques such as Western Blot, ELISA and confocal
microscopy. I am now enrolled in a graduate medicine course and was keen to bring
together my laboratory and clinical experience. I was particularly interested in exploring
how new technologies are revolutionising biomedical research and clinical practice.
New advances in biomedical research
As part of my placement at Newcastle University’s immunology lab led by Professor
Haniffa, I was introduced to cutting-edge single cell technologies. I learned that
high-dimensional analysis of single cells is providing new insights into biological
processes and pathology. Past research has focused on morphology, physical properties
and surface markers. However, this approach only facilitates study of limited cellular
parameters, which introduces intrinsic bias and can lead to erroneous conclusions.
Subsequent exploration of gene expression and function has been conducted on a population
level, taking an average from a sample of many cells. This involved prior identification
and isolation of cells using techniques such as flow cytometry, which is limited by
a finite number of parameters. Study of the transcriptome (transcriptomics), which
can be defined as the complete set of RNA transcripts in a cell under specific conditions,
has opened up further avenues for single cell research. Single cell transcriptomics
does not require a priori selection, enabling researchers to use the transcriptome of individual cells
for their identification. Thus, single cell mRNA sequencing (scRNA-seq) data can be
accrued to subsequently direct functional study with greater accuracy. As this approach
considers many more parameters, it minimises bias within the experimental system.1
Bioinformatics
A key requirement to maximise the benefits of this new research is robust bioinformatics
capabilities. Bioinformatics utilises computation to convert biological data into
interprétable information. Genomics is producing increasing quantities of data which
need to be stored, processed, interpreted and visualised. A suite of analysis programmes
is available which are statistically-based and often incorporate machine learning
algorithms.2
There is great complexity underlying the programming to manage data produced from
scRNA-seq experiments. An in-depth understanding of the computing behind informatics
is not necessary, but an ability to utilise and navigate algorithms to interpret data
is likely to become an essential part of research in the future.
Single cell RNA-sequencing in immunology
The seminal study demonstrating the utility of scRNA-seq to define immune cell types
without prior selection was performed on mouse spleen.3 More recent research in human immunology exploits scRNA-seq. I will focus on two
studies of human dendritic cells to demonstrate the underlying scRNA-seq experimental
approach and bioinformatics concepts in interpreting scRNA-seq data. Villani et al.
performed full length mRNA transcript measurement whereas See et al. measured short
sequences at the 3’ end of mRNA transcripts which were coupled to a unique molecular
identifier for each gene.4-5
Figure 1.
Human blood Dendritic Cell (DC) heterogeneity delineated by single-cell RNA sequencing.
t-SNE visualisation of scRNA-seq datasets of 742 cells. Up to five top discriminators
are listed next to each cluster. Each dot represents an individual cell. (From Figure lC Villani et al. Single-cell RNA-seq reveals new types of human blood dendritic cells,
monocytes, and progenitors. Science. 2017; 356(6335). Copyright© (2017) [The American
Association for the Advancement of Science]. Reprinted with permission from The American
Association for the Advancement of Science.
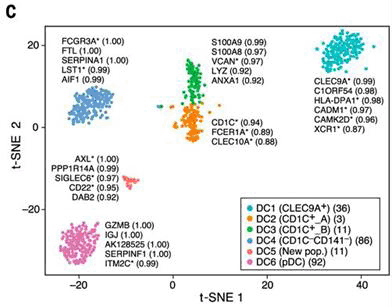
Next generation sequencing of the cDNA from single cells produces millions of short
reads which need to be aligned to the reference human genome. There are several packages
such as Seurat which provide an all-in-one application suite to undertake quality
control, analysis, exploration and visualisation of scRNA-seq data. One of the main
analyses undertaken is to cluster cells by their respective transcriptome profiles.
Cell clusters can be visually displayed using t-distributed stochastic neighbour embedding
(t-SNE), which enables high-dimensional datasets to be represented in two dimensions
(Figure 1).4
Genes that best characterise each cell cluster identified can also be represented
as a heat map, used by Villani et al. (Figure 2).4
Figure 2.
Heat map display of quantified gene expression. (Figure lD from Villani et al. Single-cell RNA-seq reveals new types of human blood dendritic
cells, monocytes, and progenitors. Science. 2017; 356(6335). Copyright© (2017) [The
American Association for the Advancement of Science]. Reprinted with permission from
The American Association for the Advancement of Science).

This displays the expression profile for the relevant discriminatory gene for each
cell (represented by each column). High intensity expression (indicated in yellow)
for the respective genes distinguish the various cell types identified from the scRNA-seq
analysis.
Further understanding of cellular heterogeneity coupled with experiments exploring
responses to various stimuli (e.g. viruses) and different cancer microenvironments
are radically changing biomedical science. Combining this powerful methodology with
proteomics and metabolomics technologies will provide unprecedented resolution to
redefine disease classification, pathogenesis and identification of novel therapeutic
targets. One existing example is immunotherapy for certain cancers, an area of research
that is already showing remarkable promise.6-8
Conclusion
This placement was a wonderful opportunity to engage first-hand with world-leading
immunological research. I was able to learn about the changing landscape of biomedical
research and the increasingly important role of bioinformatics. I would highly recommend
that fellow medical students seek out similar opportunities throughout their training
as biomedical research is a rapidly evolving field that will impact on how medicine
will be practised in the future.
Acknowledgments
I would like to acknowledge Professor Haniffa and her team for welcoming me into their
lab and for the subsequent input from Professor Haniffa to help edit and refine this
article.
Conflict of Interest Statement & Funding
The Authors have no funding, financial relationship or conflicts of interest to disclose.
Author Contributions
Conceptualization: BS. Methodology: BS. Writing - Orginal Draft: DS. Writing Review
& Editing: BS. Visualization: BS.
References
1.Liang J, Cai W, Sun Z. Single-cell sequencing technologies: current and future. J Genet Genomics. 2014 Oct 20;41(10):513–28.
2.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015 Mar;16(3):133–45.
3.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, . Massively parallel single cell RNA-Seq for marker-free decomposition of tissues into
cell types. Science. 2014 Feb 14;343(6172):776–9.
4.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, . Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and
progenitors. Science. 2017 Apr 21;356(6335).
5.See P, Dutertre CA, Chen J, Günthet P, McGovern N, Irac SE, . Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017 Jun 9:356(6342).
6.van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605.
7.Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, . Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016 Jun 16;534(7607):396–401.
8.Graciotti M, Berti C, Klok HA, Kandalaft L. The era of bioengineering: how will this affect the next generation of cancer immunotherapy? J Transi Med. 2017 Jun 19;15(1):142.
Ben Sayer, 1 St George’s, University of London, London, UK
Mihnea-Alexandru Găman, Editor
About the Author: Ben Sayer is a final year medical student at St George’s, University of London, London,
UK.
Correspondence: Ben Sayer, Address: St George’s, University of London, London, UK. Email: bensayer@hotmail.co.uk
Cite as: Sayer B. New Frontiers in Biomedical Research: A Medical Student Perspective. Int J Med Students. 2018 Sep-Dec;6(3):120-122.
Copyright © 2018 Ben Sayer
This work is licensed under a Creative Commons Attribution 4.0 International License.
International Journal of Medical Students, VOLUME 6, NUMBER 3, December 2018

